![]() inGAP-sv: structural variation detection and visualization
inGAP-sv: structural variation detection and visualization
Ji Qi (qij@fudan.edu.cn) and Fangqing Zhao (zhfq@mail.biols.ac.cn)
12/16/2010
We developed an
integrative next-generation genome analysis pipeline (inGAP), which employed a
Bayesian principle to detect single nucleotide polymorphisms (SNPs), small
insertion/deletions (indels). inGAP has been applied
to a number of genome projects, including bacteria, yeast, plants and mammals.
Here we extend this pipeline to identify and visualize large-size structural
variations, including insertions, deletions, inversions and translocations.
1.
What inGAP-sv can do?
á
Refine short
read alignment by re-aligning short reads around a putative SV.
á
Detect
large-size structural variations using paired end sequencing reads.
á
Visualize
SAM-formatted alignments and SVs.
2.
How does inGAP-sv identify SVs?
á
Classify
mapped paired-end reads into normal/anomalous mapping types.
á
Detect Ògapped
regionsÓ which cannot be covered by normally mapped paired reads.
á
Detect SVs
based on various anomalous mapping combinations. Read qualities, mapping
qualities, and ratio of paired-end reads relative to average mapping densities
will be considered for the calculation of SV quality.
3.
How to get started?
á
inGAP-sv
requires two files, a FASTA formatted reference sequence and a SAM alignment
á
A PTT
formatted annotation file for the reference sequence is optional.
á
A demo
application is preloaded in inGAP-sv.
4.
WhatÕs the difference between
inGAP-sv and other SV tools?
á
Most of the
current SV tools can only detect very short indels (e.g. 1-10bp); inGAP-sv and
a few others (e.g. breakdancer) work well with large SVs (>100bp)
á
inGAP-sv is a one-stop SV detector. Users can identify,
visualize, annotate and manually edit SVs using inGAP-sv.
á
Compared with
other command-line based SV tools, visualization of paired reads in inGAP-sv
can significantly reduce the false discovery rate.
5.
WhatÕs the performance of inGAP-sv?
á
We firstly
tested inGAP-sv using simulated data with large SVs (100-1000bp) from the
Yoruban genome (NA18507). inGAP-sv could successfully
identify 75%-90% of large indels and >85% of inversions with high accuracy
rate. Detailed evaluation is in progress.
á
We also
applied inGAP-sv to an Arabidopsis
thaliana genome re-sequencing project. inGAP-sv
have identified 815 insertions and 1000 deletions. We compared these indels to
the Monsanto A. thaliana assembly,
and found that 78% of the deletions could be covered by Monsanto contigs and
99% of them were correct. 71% of insertions could be covered by Monsanto
contigs and 96% of them were correct.
á
inGAP-sv supports parallel computing.
6.
How to access inGAP-sv?
á
Users can
download the latest version of inGAP from http://sourceforge.net/projects/ingap/.
We provide binaries for Windows, Linux, MacOS/X.
á
A quick manual
is available at http://schuster-33.bx.psu.edu/shared/manual.pdf .
7.
Screenshots
á
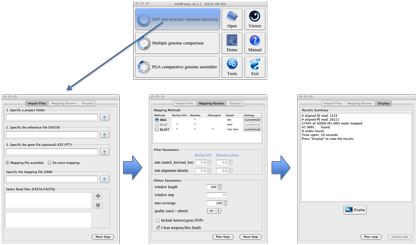
Main functions
and work flow of inGAP-sv
á
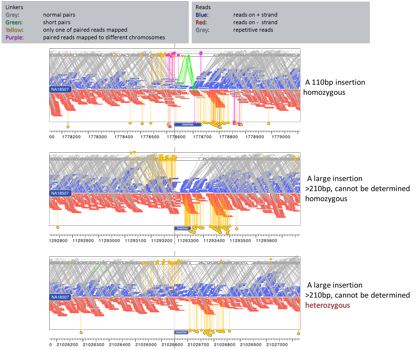
Deletions
detected by inGAP-sv
á
Insertions
detected by inGAP-sv
á
Inversions
detected by inGAP-sv
á
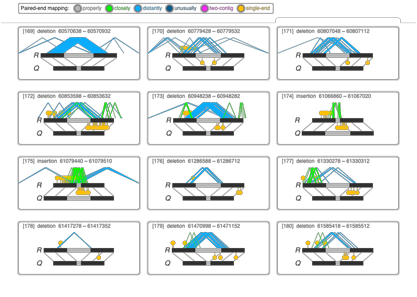
Schematic view
of SVs